
Integrations















AutoNetCan Pipeline
A comprehensive workflow for constructing biomolecular networks for translational cancer systems biology
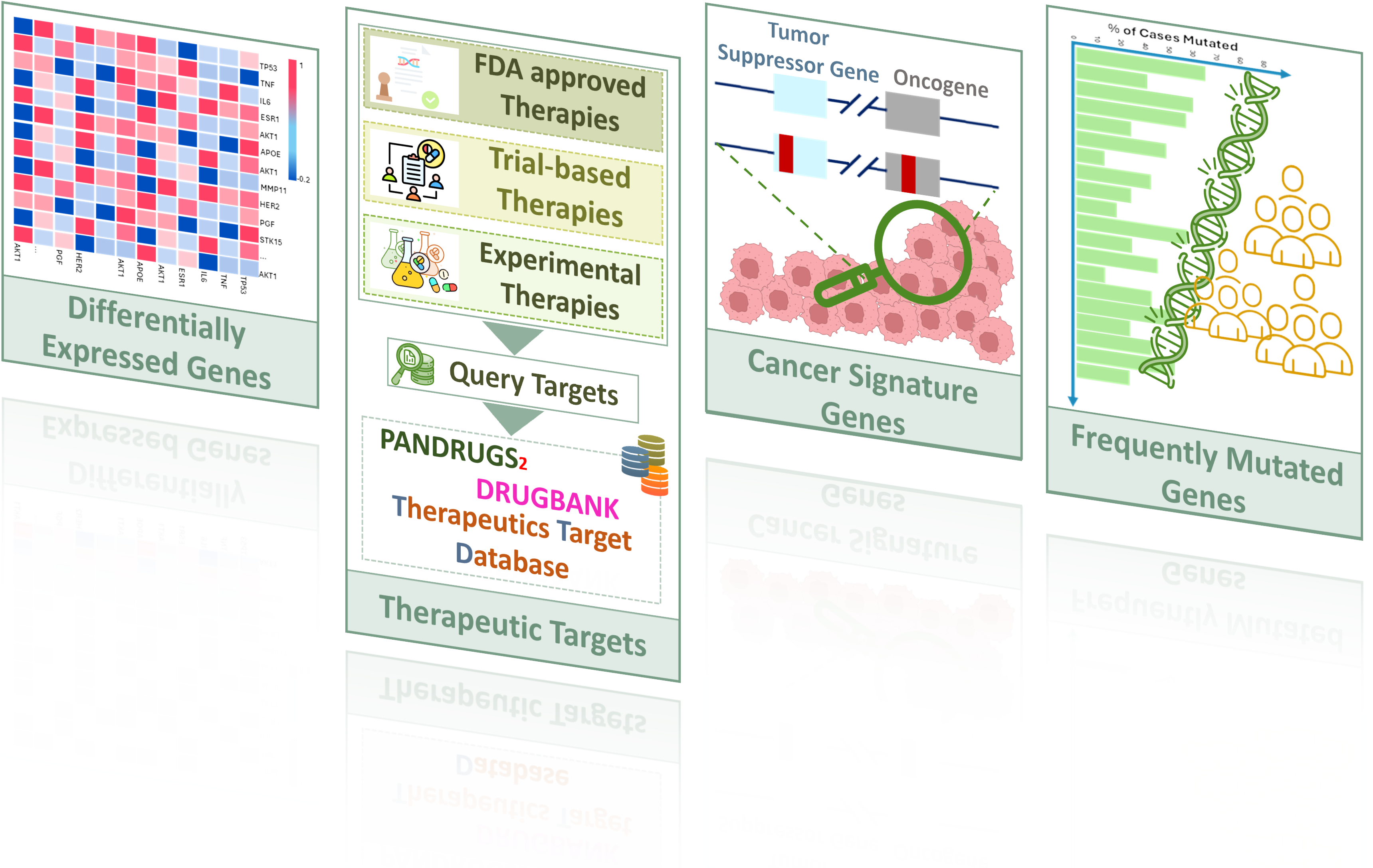
Acquisition of Nodes
The development of a biomolecular cancer model starts with the acquisition of nodes from diverse multiscale sources. We integrate omics datasets from Genomic Data Commons (GDC), frequently mutated nodes, cancer signature genes, and therapeutic targets to ensure biological relevance and translational potential.
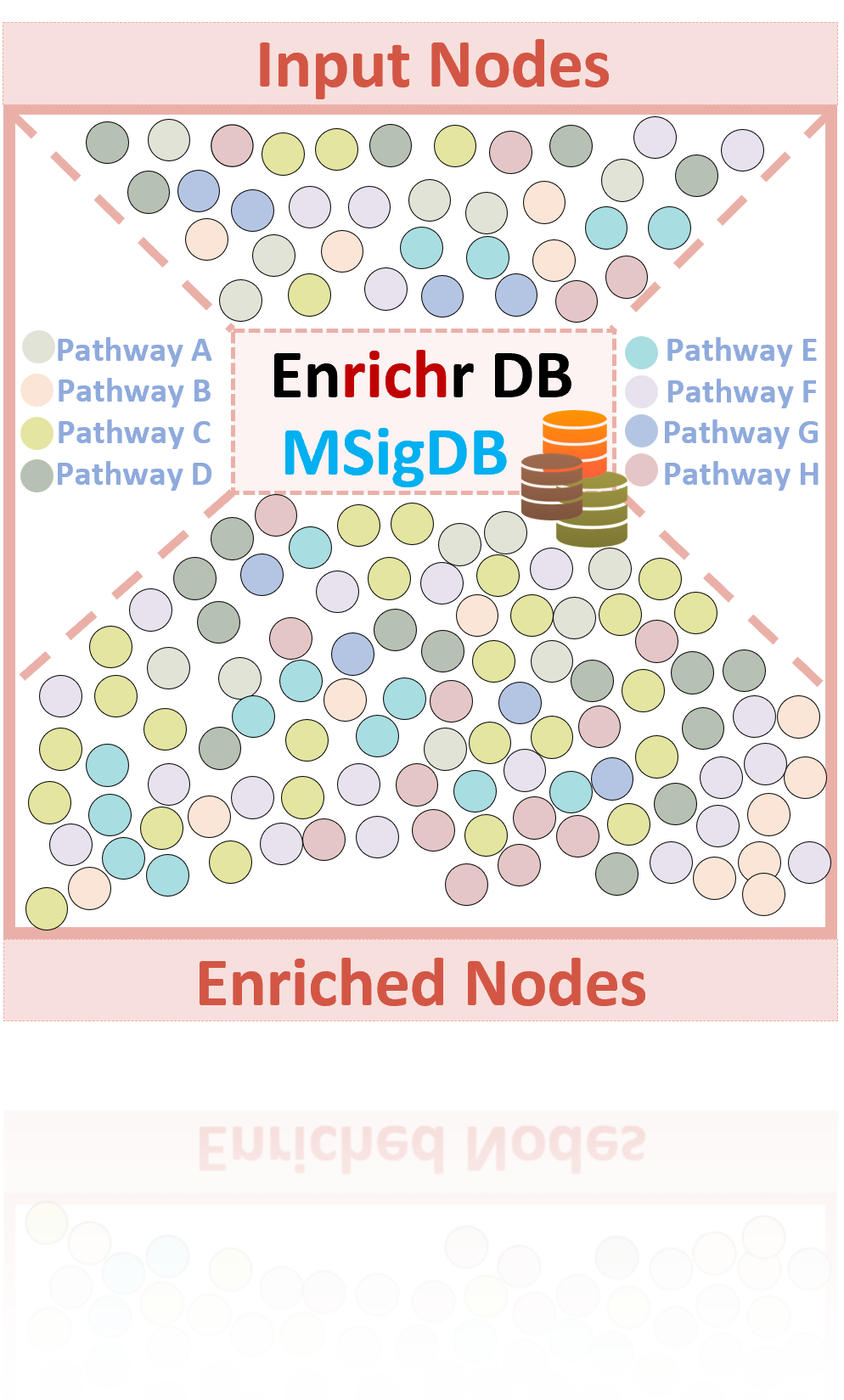
Node Enrichment
Node enrichment is performed by leveraging Enrichr, MSigDB, and curated pathway databases to represent molecular functions, cellular compartmentalization, and biological processes. This enrichment populates the network model with key interactions and pathways essential for modelling cancer biology.
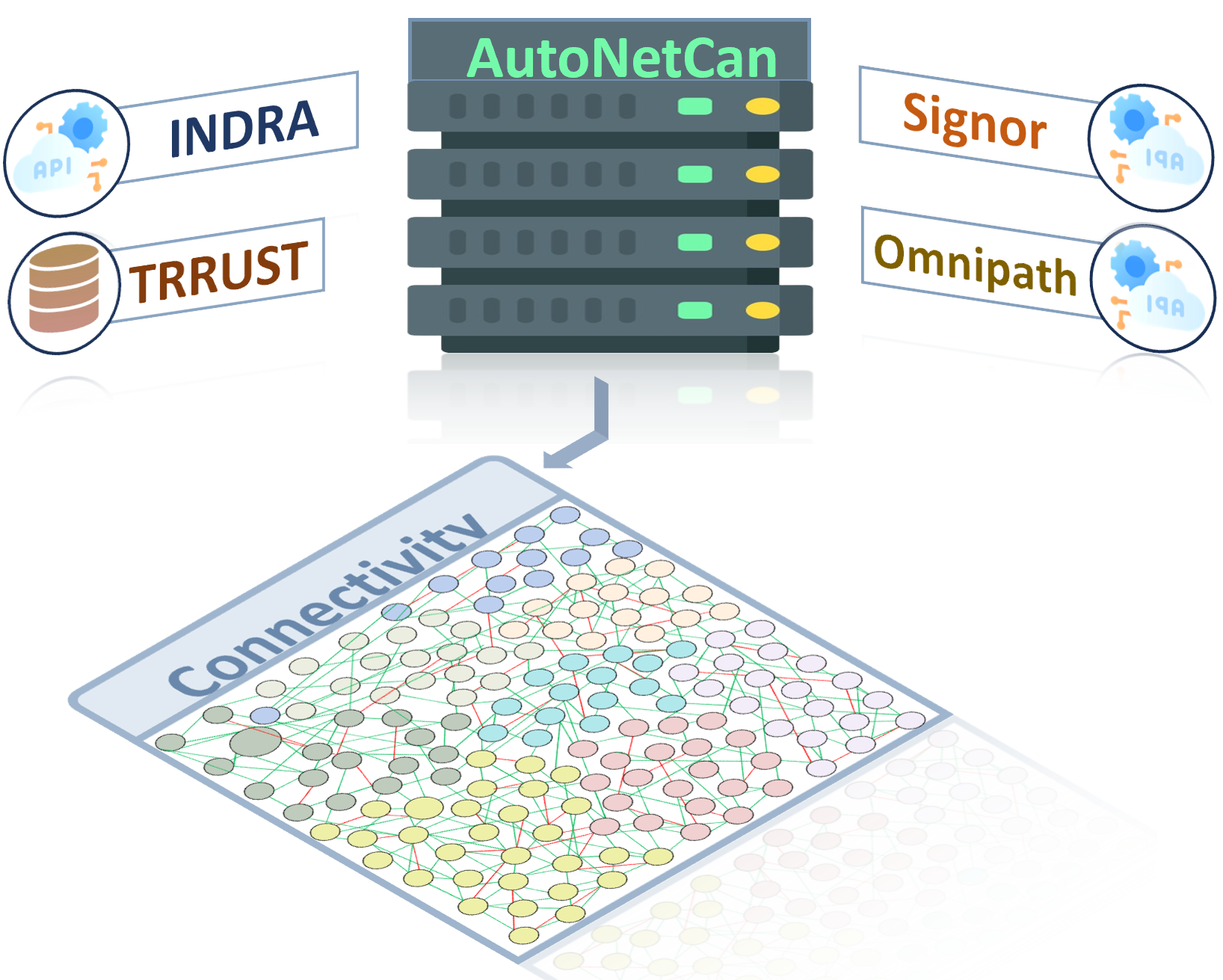
Connecting Maps & Interactome
The enriched node set is integrated into a comprehensive interactome using public databases such as INDRA, TRRUST, SIGNOR, and Omnipath. These resources inform activations and inhibitions, enabling the construction of a biologically informed network that maps essential molecular interactions and regulatory mechanisms.
Logical Modeling
Using RNA sequencing data and interactome maps, we annotate network nodes to construct optimal Boolean network models. These models enable simulation and analysis, supporting downstream applications in cancer research and therapeutic discovery.

In-Silico Cancer Models
The resulting models capture the regulatory logic of tumor-specific networks and serve as scaffolds for downstream data annotation and analysis. These models can be integrated with external tools to explore patient-specific behaviors, therapeutic responses, and personalized treatment strategies—advancing translational applications in precision oncology.

Publications
Our research contributions to cancer systems biology and personalized therapeutics
Atlantis - Attractor Landscape Analysis Toolbox for Cell Fate Discovery and Reprogramming
Scientific Reports - Nature
Read Publication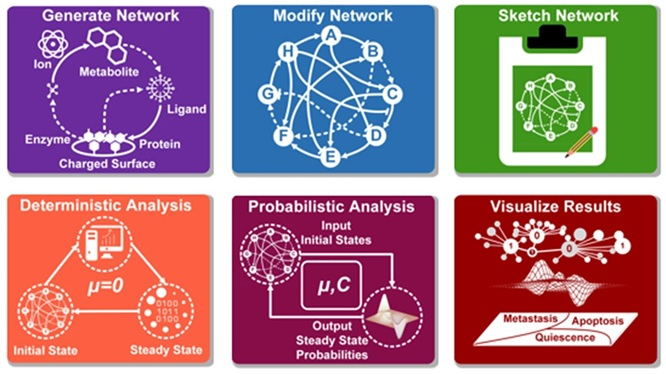
Navigating Multi-scale Cancer Systems Biology towards Model-driven Personalized Therapeutics
Frontiers in Oncology - Special Issue on "Combinatorial Approaches for Cancer Treatment"
Read Publication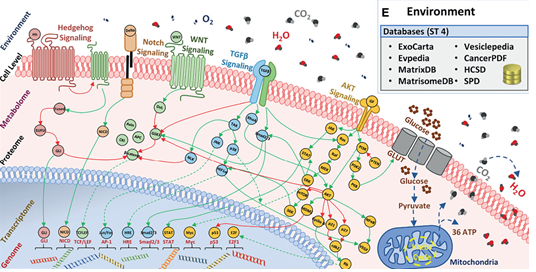
A Personalized Therapeutics Approach Using an In silico Drosophila Patient Model Reveals Optimal Chemo- and Targeted Therapy Combinations for Colorectal Cancer
Frontiers in Oncology - Special Issue on "Combinatorial Approaches for Cancer Treatment"
Read Publication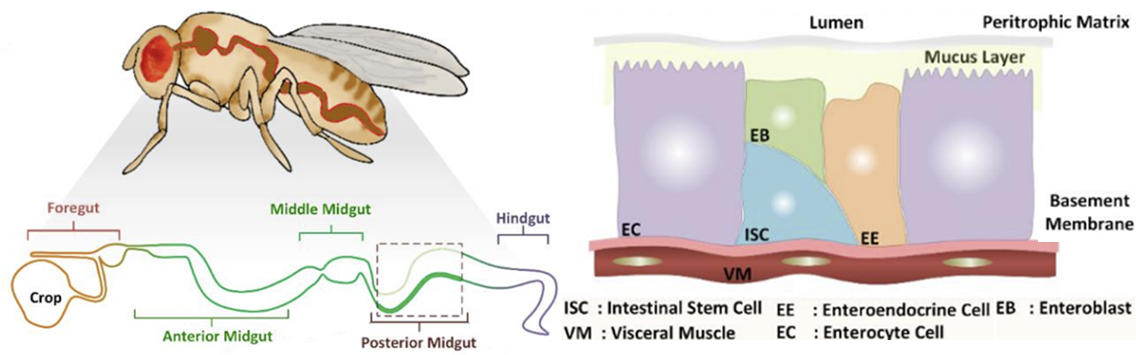
CanSeer: A Method for Development and Clinical Translation of Personalized Cancer Therapeutics
bioRxiv - the preprint server for Biology
Read Publication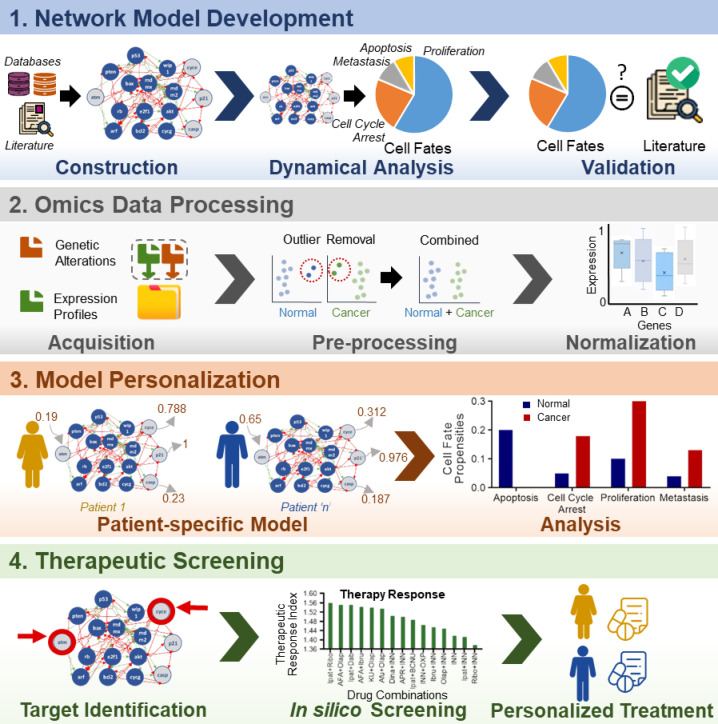
Help & Resources
Get started with AutoNetCan using our comprehensive documentation and video tutorials
Video Tutorials
Tutorial 1: Breast Cancer Network
A walkthrough of breast cancer network construction
Tutorial 2: Prostate Cancer Network
A walkthrough of prostate cancer network construction
Tutorial 3: Cytoscape Visualization
Network construction and visualization in Cytoscape
Tutorial 4: TISON Analysis
Network construction with visualization and analysis in TISON
Tutorial 5: Cosmograph Visualization
Network visualization using Cosmograph
Contact Us
Get in touch with our team for questions, support, or collaboration opportunities

Research Laboratory
Biomedical Informatics and Engineering Research Laboratory (BIRL)
Department of Life Sciences, School of Science and Engineering
Lahore University of Management Sciences (LUMS)
Lahore, Pakistan
Send us a Message
Fill out the form below and we'll get back to you as soon as possible



